

To maximize ease of use, the pipeline incorporates all the pre-requisite tools baked into a Docker image along with custom Motifizer scripts required for data processing.
HOMER ANNOTATE PEAKS SOFTWARE
We developed the Motifizer software with an aim to make it independent of the computational infrastructure and expertise. Taken together, we provide a versatile tool which can be employed by the larger user base for parsing of sequencing datasets and motif-based analysis of genomic sequences. In addition, we introduce an Analysis module which uses the employs the FIMO software of Meme Suite to analyze and compare TF motif occupancy information in user provided genomic sequences. While the parameters used for alignment and processing of bam files are set to default, the shell scripts for such processes can be easily changed to incorporate changes as required by the user. In this context, we have developed an easy to use tool which when deployed by the user using a docker container on a Linux platform, installs the various open source packages required for analysis of ChIP-seq and RNA-seq data. In addition, while individual programs exist to calculate the occurrence and/or presence of transcription factors ( Grant, Bailey and Noble, 2011), we could not find a software which could quantify and compare TF occupancy information between two or three given set of sequences. While certain programs exist which provide a detailed analysis pipeline for ChIP and RNA seq datasets ( Liu et al., 2011 Afgan et al., 2018 Batut et al., 2021), we wanted to build a software which will be fairly simple in terms of initial analysis, and therefore, more accessible to the user base with no programming expertise. However, most of these tools require some prior knowledge of programming language or computational dependencies and requirements. The analysis of ChIP-seq and RNA-seq data involves various steps performed by individual programs ( Zhang et al., 2008 Li and Durbin, 2009 Li et al., 2009, 2009 Robinson, McCarthy and Smyth, 2010 Anders, Pyl and Huber, 2015 Kim et al., 2019). Overlaying chromatin occupancy information of a TF with the regulation status of its target genes, combined with the precise location and abundance of the TF in target enhancer sequences, not only provides the gene regulatory network of the TF, but also sheds critical insights into the mechanisms of transcriptional regulation. On the other hand, the correlation of chromatin occupancy to the regulation of a target gene by a TF comes from differential gene expression (DGE) analysis performed on transcriptomic data (RNA-seq) (reviewed in Wang, Gerstein and Snyder, 2009). Chromatin occupancy profile of a TF is obtained by fixing proteins onto the DNA using a chemical agent and selectively purifying the chromatin using antibodies followed by sequencing (ChIP-seq)(reviewed in Park, 2009). Characterization of GRN of a TF in a given biological context involves two major steps: 1) Identifying the chromatin occupancy of the TF in the genome 2) Correlation of the chromatin occupancy to the regulation of its target genes.
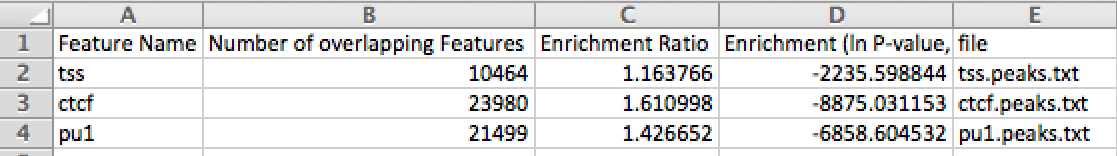
Since GRNs are critical in development and differentiation (and in disease research), identifying target genes of a TF, or the TF targeting a given gene, is important to understand the basis of all biological processes.

The complete repertoire of interactions between a particular transcription factor (TF) with its target genes constitute a gene regulatory network (GRN).


 0 kommentar(er)
0 kommentar(er)
